About
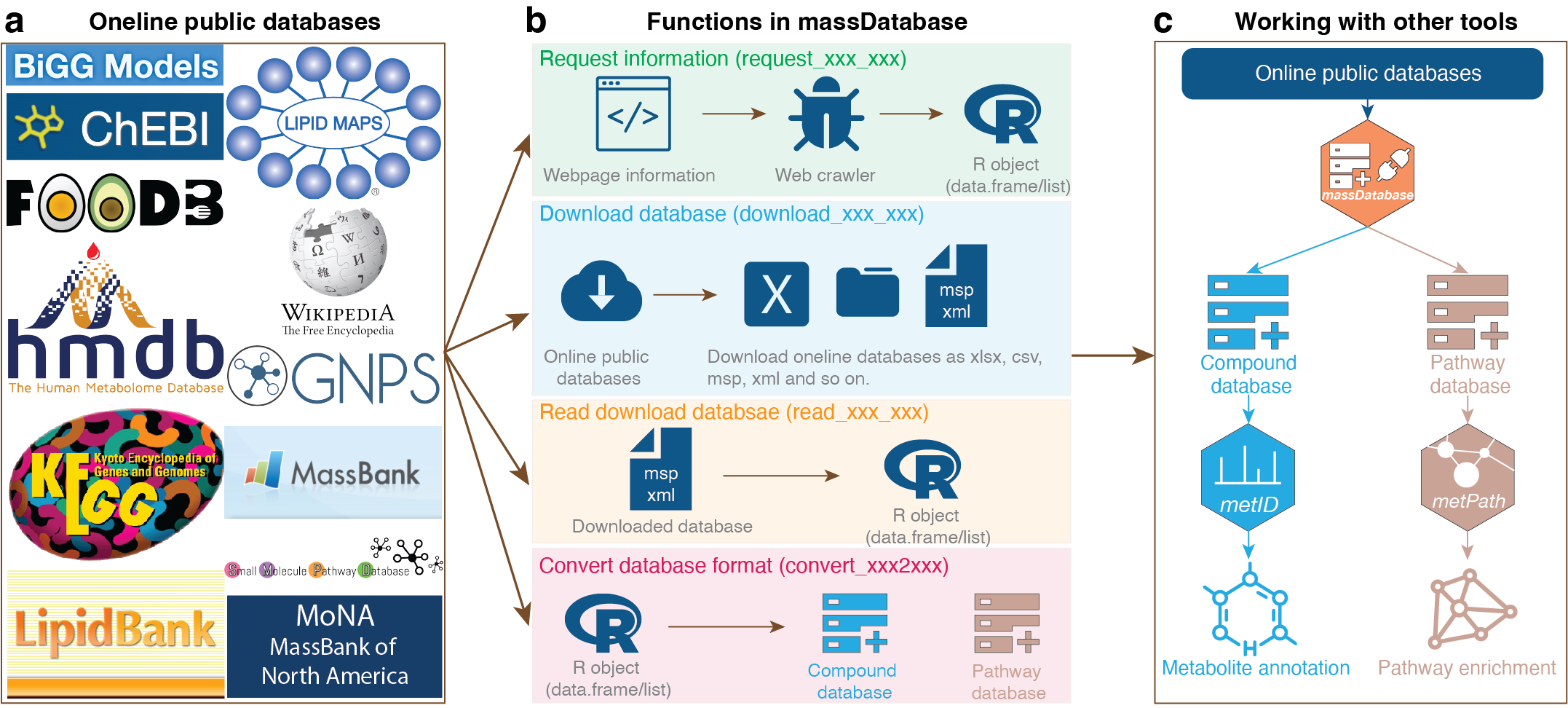
massdatabase is an R package that operates the online public databases and combines with other tools for streamlined compound annotation and pathway enrichment analysis. massDatabase is a flexible, simple, and powerful tool that can be installed on all platforms, allowing the users to leverage all the online public databases for biological function mining.
Installation
You can install massdatabase from GitLab
if(!require(remotes)){
install.packages("remotes")
}
remotes::install_gitlab("jaspershen/massdatabase")or Github
remotes::install_github("tidymass/massdatabase")Usage
Please see the Help documents page to get the instruction of massDatabase.
Need help?
If you have any questions about massdatabase, please don’t hesitate to email me (shenxt@stanford.edu).
Citation
If you use massdatabase in your publications, please cite this paper:
massDatabase: utilities for the operation of the public compound and pathway database. Web Link
Shen, X., Yan, H., Wang, C. et al. TidyMass an object-oriented reproducible analysis framework for LC–MS data. Nat Commun 13, 4365 (2022). Web Link.
Thanks very much!
